Introduction¶
OntoMerger is an ontology alignment library for deduplicating knowledge graph nodes, (i.e. ontology concepts) that represent the same domain, e.g. diseases, and connecting them to form a single directed acyclic hierarchical graph (DAG) (i.e. an ontology class hierarchy). The library implements a pipeline that takes nodes, mappings and (disconnected) hierarchies as input and produces node merges and a connected hierarchy. It also provides analysis and data testing for fine tuning the inputs in order to further reduce duplication, as well as to increase connectivity.
>@article{ontomerger,
arxivId = {???},
author = {Geleta, David and Rozemberczki, Benedek
and Nikolov, Andriy and O'Donoghue, Mark
and Gogleva, Anna and Tamma, Valentina},
month = {may},
title = {{OntoMerger: An Ontology Alignment Library for
Creating Minimal and Connected Domain Knowledge
Sub-graphs.}},
url = {http://arxiv.org/abs/???},
year = {2022}
}
Overview¶
Motivation¶
This library is used in production at the Biological Insights Knowledge Graph project (AstraZeneca). Throughout this document, we will use real life examples and statistics from this project. For more details about BIKG, please read the paper.
Knowledge fragmentation¶
Building knowledge graphs, especially in the biomedical domain, often involves integrating data from several overlapping but heterogeneous data sets. One data set may provide information about genes, another about diseases, and a third about how certain genes are related to a given disease. Different data sets might use different vocabularies (ontologies, taxanomies etc.) to refer to the same concepts, causing syntactic and semantic heterogenity in the integrated data set.
Heterogenity leads to fragmented knowledge that hinders the efficacy of knowledge graphs: we might know what genes are causing the a bronchial disease asthma, and which drugs can be used for treatment, but if these two statements (triples) are using different identifiers, a recommender system would not pick up this pattern and suggest using the same drug for other related diseases.
- Example of knowledge fragmentation (by merge the two asthma node instances into
a single node, the two separate statements become connected):
GENE -- participates --> DOID:2841 (Asthma)
MONDO:0004979 (Asthma) <-- treats -- COMPOUND
Example of resolving knowledge fragmentation:
DOID:2841 -- merge --> MONDO:0004979
GENE -- participates --> MONDO:0004979 (Asthma) <-- treats -- COMPOUND
Node duplication¶
The BIKG data set contains approximately 189k Disease nodes from over 16 different sources. The figure below shows the count and frequency of the top ten Disease sources.
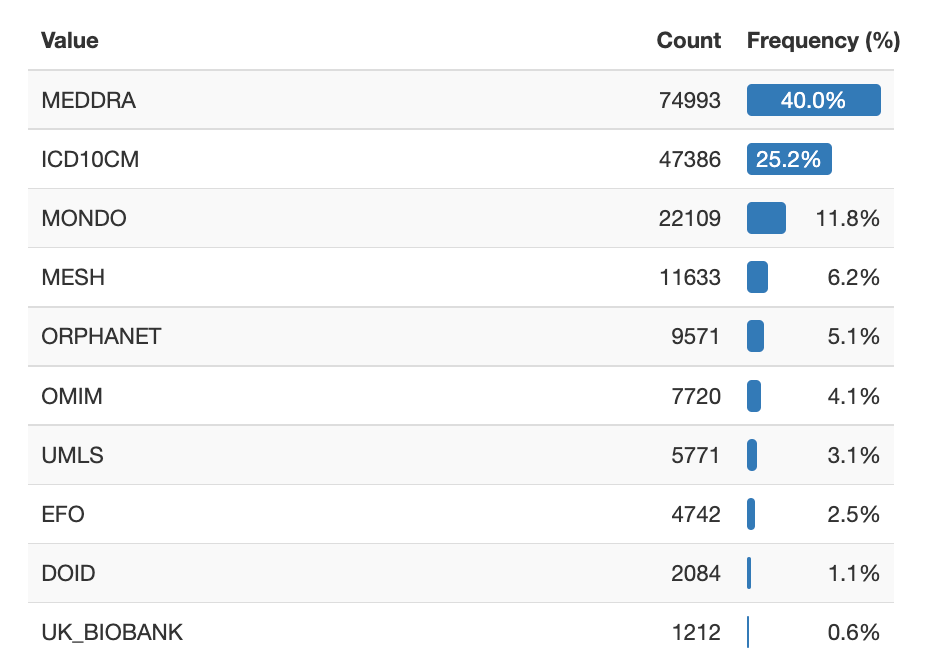
Frequency chart of the top ten Disease node sources in the example data set.¶
Data set version discrepancies¶
Data set version discrepancy can also lead to node duplication. Data sets may be stale or updated with varying frequency. Even if two data set use the same source they might use different versions. This can also cause issues as concepts can be deprecated: in a simple case the a deprecated concept can get another identifier (one to one internal code reassignment). In a more complex case, a concept may be removed, or split (refined) into multiple concepts.
Disconnected nodes¶
Organising domain nodes into a single hierarchy improves ML processes (pattern recognition). Hierarchies facilitate finding similar nodes based on neighborhoods (e.g. sibling nodes). Some of the challenges presented by heterogeneous data sets are that
- not all nodes have (available) hierarchies (this could be due to licensingissues, dynamic data sets such as BIKG, etc.);
- hierarchies may be incompatible (e.g. ICD10CM is not a proper hierarchy,whereas MONDO is);
typically not all nodes can be aligned to the same vocabulary.
As a result, without any intervention, we are left with several vocabulary fragments, i.e. not a single DAG that properly supports the aforementioned ML; or an inconsistent hierarchy (loops etc >> todo include refs.).
Method¶
The goals of the alignment process are to (1) reduce node duplication of domain nodes, and (2) to connect the domain nodes. The library does not aim to create a merged ontology that retains all knowledge from the integrated parts (i.e. all hierarchy connections, and the complete granularity of the nodes). Therefore the methods we employ are approximations that make use of the available background knowledge to achieve these goals (availability of background knowledge is an issue in dynamic settings, especially regarding mappings, that are often produced and curated by humans).
Scaffolding seed ontology, all are merged onto it and connected to it. Secondary hierarchies that we try to preserve to some degree.
Using Pandas
Providing analysis and data tests
Running the project
$ onto_merger --f PROJECT_FOLDER